Gastrointestinal System Infection Modeling & Pharmacodynamics Services
Introduction
Gastrointestinal System infection diseases, such as viral gastroenteritis, bacterial Dysentery, and parasitic infections, caused by various pathogens, including Norovirus, Salmonella species, Shigella species, Escherichia coli (ETEC/EHEC), Clostridioides difficile (CDI), and Giardia lamblia, represent a significant global public health burden. These infections are among the leading causes of morbidity worldwide and are responsible for millions of hospitalizations and hundreds of thousands of deaths annually, particularly in children under five. The impact is particularly pronounced in low-resource settings with poor sanitation. Creative Biolabs provides a portfolio of specialized animal models that accurately replicate the clinical features of digestive system infectious diseases. These models can be used for the pharmacological and pharmacodynamic evaluation of oral antibiotics, antiviral drugs, enterotoxin neutralizers, microbiota therapies (FMT/probiotics), and oral vaccines, thereby accelerating the new drug development process.
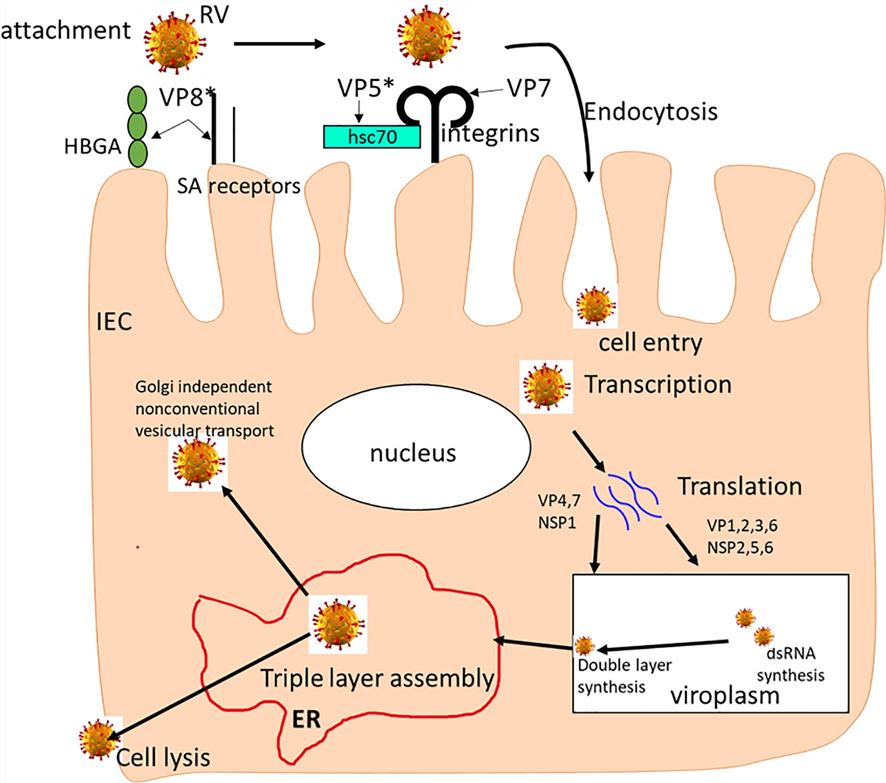 Fig.1 The rotavirus replication cycle.1
Fig.1 The rotavirus replication cycle.1
Available Gastrointestinal System Infection Models
Leveraging over a decade of dedicated expertise in infectious disease modeling, Creative Biolabs offers a robust array of well-characterized and clinically relevant digestive system infection animal models. These advanced models are designed to expedite your R&D process, ensuring efficient and reliable preclinical evaluation.
| Gastrointestinal System Infection Models | RelatedDisease & Drug Evaluation | Animal Species |
| Vibrio cholerae Infection Models | Cholera, Acute Secretory Diarrhea; ideally suited for therigorous evaluation of Antibiotics (e.g., Tetracycline, Azithromycin), ORS, andVaccines. | Mouse,Rat,Rabbit, NHPs |
| Shigella spp. Infection Models | Bacillary Dysentery (Shigellosis), Invasive Diarrhea;highly effective for the assessment of Antibiotics (e.g.,Azithromycin), Vaccines, and Anti-virulence agents. | Guinea Pig, Rabbit, Mouse, Rabbit |
| Typhoid andParatyphoid Fever Infection Models | Typhoid/Paratyphoid Fever, Systemic Enteric Fever;specifically engineered for the evaluation of Antibiotics (e.g.,Fluoroquinolones, Azithromycin), and Vaccines. | Mouse, Rat, Bamaminiature pig, NHPs |
| Helicobacter pyloriInfection Models | Chronic Gastritis, Peptic Ulcer Disease, Gastric Cancer;very suitable for the comprehensive evaluation of Combination AntibioticTherapy (e.g.,Triple Therapy), and Vaccines. | Mouse, Rat, Gerbil, Guinea Pig, Pig |
| Enterohemorrhagic Escherichia coli (EHEC)Infection Models | Hemorrhagic Colitis, Hemolytic Uremic Syndrome (HUS);ideally suited for the rigorous evaluation of Supportive care, Shiga ToxinNeutralizing Agents, and Anti-adhesion therapies. | Mouse, Rabbit, Pig, Ferret, NHPs |
| Clostridioides difficile Infection (CDI) Models | C. difficile-Associated Diarrhea (CDAD), Recurrent CDI; highlyeffective for the assessment of Antibiotics (e.g., Fidaxomicin), FMT, Toxin neutralizingantibodies, and Vaccines. | Mouse, Hamster, Guinea Pig, Rat |
| RotavirusGastroenteritis Infection Models | Acute Gastroenteritis (severe diarrhea in children);specifically engineered for the evaluation of Vaccines (e.g., Liveattenuated), Antivirals, and Probiotics. | Mouse, Pig, Rabbit |
| Norovirus InfectionModels | Epidemic Acute Gastroenteritis (Winter Vomiting Disease);very suitable for the comprehensive evaluation of Supportive care, Antivirals(e.g.,Nucleoside analogs), and Vaccines. | Mouse, Rat,NHPs, Piglet, Calve |
| Hepatitis E Virus(HEV) Infection Models | Hepatitis E (Acute/Chronic), Fulminant Hepatic Failure;ideally suited for the rigorous evaluation of Antivirals (e.g., Ribavirin)and Vaccines. | Mouse, Mongolian Gerbil, Rat, Rabbit, Pig, NHPs |
| AdenovirusGastroenteritis Infection Models | Acute Gastroenteritis (e.g., Type 40/41); highly effective for theassessment of Supportive care and Antivirals (e.g., Cidofovir inimmunocompromised). | Mouse, Rat, Rabbit, Pig, NHPs,Dog |
| Entamoeba histolytica Infection Models | Amoebiasis, Amoebic Dysentery, Amoebic Liver Abscess;specifically engineered for the evaluation of Antiprotozoal drugs (e.g.,Metronidazole) and Luminal agents. | Mouse, Rat, Mongolian Gerbil, Rabbit |
| Giardia lamblia Infection Models | Giardiasis (Beaver Fever), Chronic Diarrhea; verysuitable for the comprehensive evaluation of Antiprotozoal drugs (e.g.,Metronidazole, Tinidazole), Nitazoxanide. | Mouse, Mongolian Jird |
| Ascaris lumbricoides (Ascariasis) Infection Models | Ascariasis, Intestinal obstruction; ideally suited forthe rigorous evaluation of Antihelminthic drugs (e.g., Albendazole, Mebendazole,Ivermectin). | Mouse,Rat,Guinea Pig, Hamster, Pig |
| Taenia spp.(Intestinal Taeniasis) Infection Models | Taeniasis (Tapeworm infection), Cysticercosis; highlyeffective for the assessment of Antihelminthic drugs (e.g., Praziquantel,Niclosamide, Albendazole). | Mouse, Rat, Pig |
| Hookworm InfectionModels | Hookworm Disease, Iron Deficiency Anemia; specificallyengineered for the evaluation of Antihelminthic drugs (e.g., Albendazole,Mebendazole), and Vaccines. | Mouse, Rat, Dog, Hamster |
| Cryptosporidium spp. Infection Models | Cryptosporidiosis, Persistent Diarrhea; very suitable forthe comprehensive evaluation of Antiprotozoal drugs (e.g.,Nitazoxanide), Supportive care. | Mouse, Rabbit, NHPs,Chicken |
| Microsporidia spp. Infection Models | Microsporidiosis (often severe in HIV/AIDS patients);ideally suited for the rigorous evaluation of Antifungal/Antiprotozoal drugs(e.g.,Albendazole, Fumagillin). | Mouse, Rat, Rabbit |
Measurements
Creative Biolabs assesses gastrointestinal system infection animal models by integrating an array of multi-dimensional technologies to comprehensively analyze disease mechanisms and drug efficacy, including but not limited to:
- Clinical Observation: Detailed recording of the animal models' symptoms (e.g., diarrhea, weight loss, dehydration, and body temperature changes).
- Pathogen Load and Identification: Accurate quantification of the pathogen load in feces, intestinal tissue, and blood is performed using quantitative PCR, ELISA, or viral culture and selective culture media. Strain identification is carried out using molecular subtyping.
- Histopathological Examination & Barrier Function: Focus is placed on inflammatory infiltration, ulceration, and tissue necrosis. Key assessment indicators include mucosal epithelial damage scoring and immunohistochemical analysis of Tight Junction proteins to evaluate intestinal barrier function.
- Molecular mechanism research: The expression profiles of inflammation-related genes are detected by RT-qPCR and RNA sequencing, to analyze the signal pathway activation mechanisms.
- Others: Intestinal microbiome analysis (16S rRNA / Metagenomics), metabolomics, and comprehensive mucosal immune analysis (e.g., Secretory IgA, lymphocyte subsets), among others.
Applications
- Disease Modeling: The models are used to simulate key pathological processes such as pathogen colonization, toxin release, epithelial damage, and microbiota dysbiosis within the gastrointestinal tract.
- Drug Discovery and Development: The models serve as essential platforms for screening oral antibiotics, antivirals, antitoxin agents, and evaluating the mucosal immune efficacy of oral vaccines.
- Therapeutic Strategy Optimization: The models are critical for validating the role of non-antibiotic interventions, such as Fecal Microbiota Transplantation (FMT), probiotics, and anti-virulence factors, in restoring gut microbiota and treating recurrent infections.
- Mechanism Study: The models dissect the multifaceted pathophysiology of enteric infections by reconstructing pathogen adhesion and invasion across the mucus-epithelial barrier, elucidating the molecular disruption of intercellular tight junctions, and characterizing the reciprocal interactions between the commensal microbiome, pathogenic agents, and mucosal immune defenses.
Our Advantages
- Comprehensive Model Library with Extensive Technical Expertise: Our library supports a wide variety of experimental animals, including mice, rats, dogs, and pigs, and can be customized to accommodate different pathogens and research requirements.
- Accurately simulate the disease process: We meticulously control the disease stage by regulating infection dose, cycle, and environmental factors to ensure precise simulation.
- Standardized Procedures and High-Quality Data Validation: A stringent model evaluation system is established for etiology verification, pathological assessment, and functional testing. Standardized modeling procedures (e.g., fasting before Helicobacter pylori infection) ensure consistency across multiple experimental batches, meeting the reliability requirements for drug screening and mechanistic research.
- Multi-dimensional technical platform support: We integrate platforms in molecular biology, immunology, virology, and pathology to provide comprehensive pharmacodynamic evaluations.
- One-Stop Service and Resource Integration: We offer customized, integrated solutions by flexibly adjusting model strategies based on drug type (e.g., small molecules, biologics) and research stage (e.g., target validation, safety evaluation). Our seamless service integrates all resources, from model development and PK/PD analysis to professional reporting, to accelerate preclinical development and eliminate vendor complexity.
Work with Us
- Summarize the project requirements and fill in the information collection form.
- Sign a CDA from both parties to further communicate information, such as targets.
- Select an animal model, discuss experimental design, and determine assay parameters.
- Project costing and project schedule forecasting.
- We provide a detailed project plan, including the required sample quantities, methods, and protocols.
- Both parties confirm the project details and start the project.
- Confirm the timeline of the project.
- We provide periodic results and information on the animal's condition.
- We will work together to make project adjustments as necessary.
- We provide a comprehensive project report promptly.
- We arrange transportation for the produced samples.
- We provide a discussion of the project results and help to arrange the next steps.
- Data storage and archiving.
FAQs
-
Q: Why are in vivo GI models essential for testing new antimicrobials?
A: The GI tract presents unique challenges (such as low pH, enzyme activity, and complex microbiota). In vivo models are essential tools for assessing the local stability of oral drugs, their bioavailability within the intestinal lumen, and their efficiency of transmucosal absorption, providing data that is more predictive than in vitro testing.
-
Q: Which specialized models do you use for Pharmacodynamic (PD) assessment in the GI tract?
A: We utilize validated models such as the CDI hamster/mouse model, Salmonella/Shigella enteric fever models, and viral enteritis models. These platforms enable precise measurement of pathogen shedding, toxin levels, gut pathology (e.g., histology), and microbiota composition under drug challenge.
-
Q: How do you assess the impact of a new drug on the gut microbiome?
A: We combine in vivo drug challenge with 16S rRNA gene sequencing and metagenomics analysis. This allows for quantitative assessment of the drug's effect on microbial diversity, specific bacterial populations, and the overall dysbiosis index, which is key for developing microbiota-sparing antibiotics.
-
Q: How does PD modeling accelerate the development of CDI therapeutics?
A: For CDI, our PD modeling focuses on the relationship between drug exposure and toxin production and disease recurrence. We determine the optimal dose by combining 16S rRNA/metagenomic sequencing. This optimal dose is capable of maximizing bacterial clearance while minimizing disruption to the protective commensal microbiota, thereby reducing the risk of recurrence and accelerating clinical readiness.
-
Q: What commercial advantages do clients gain from your GI PD services?
A: Our scientifically rigorous, in vivo-driven data package reduces attrition in later development stages by validating efficacy in a complex biological environment. This provides the data confidence necessary to attract investment and support compelling regulatory submissions for treating critical GI infections.
Published Data
The memory humoral immunity induced by C. difficile infection (CDI) is only targeted against the virulence toxins (TcdA/TcdB). Secondary infection significantly reinforces this IgG response, but shows no memory activation against surface proteins (SlpA/FliC), and the immunodominance of TcdA remains unchanged throughout. This finding provides a crucial direction for CDI vaccine development.
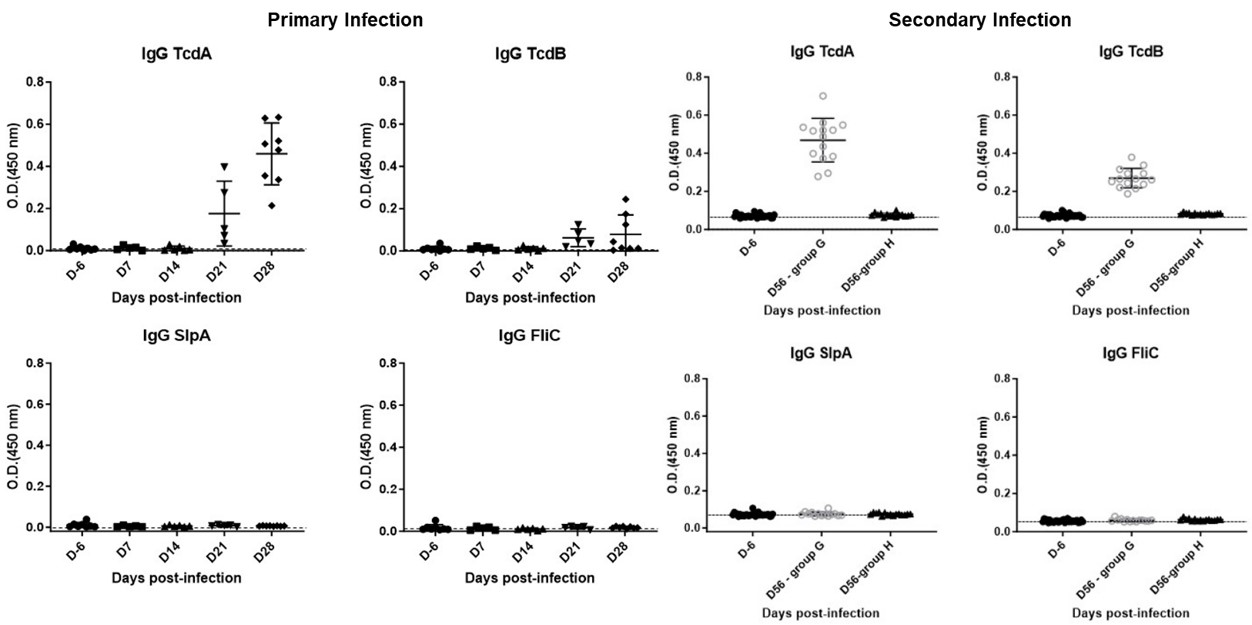 Fig.2 Serum IgG kinetics following primary and secondary infections.2
Fig.2 Serum IgG kinetics following primary and secondary infections.2
References
- Amimo, Joshua Oluoch et al. "Rotavirus Interactions With Host Intestinal Epithelial Cells." Frontiers in immunology vol. 12 793841. https://doi.org/10.3389/fimmu.2021.793841. Distributed under Open Access license CC BY 4.0, without modification.
- Mizrahi, Assaf et al. "A Mouse Model of Mild Clostridioides difficile Infection for the Characterization of Natural Immune Responses." Microorganisms vol. 12,10 1933. https://doi.org/10.3390/microorganisms12101933. Distributed under Open Access license CC BY 4.0, with modification.
For Research Use Only.
