Antimicrobial Resistance (AMR) Modeling & Pharmacodynamics Services
Introduction
Global public health faces an urgent crisis driven by Antimicrobial Resistance (AMR), where the rapid spread of resistant strains leads to sharply increased treatment failure rates and mortality. Crucially, for critically ill and immunocompromised patients, infections by Multidrug-Resistant (MDR) pathogens often render medical intervention ineffective, posing a direct threat to life. Furthermore, AMR forces routine medical procedures like surgery and chemotherapy to carry prohibitively high infection risks. Creative Biolabs specializes in designing highly customized Pharmacodynamic (PD) models, focusing on resolving the critical challenges of effective drug penetration into infection sites and determining the optimal drug exposure required to overcome resistant strains. We also utilize resistant bacterial animal models as an indispensable bridge between in vitro studies and clinical validation, enabling the comprehensive assessment of the in vivo efficacy of novel therapeutics, including new antibiotics, biologics, phage therapy, and anti-virulence drugs, thereby accelerating the pipeline for combating drug resistance.
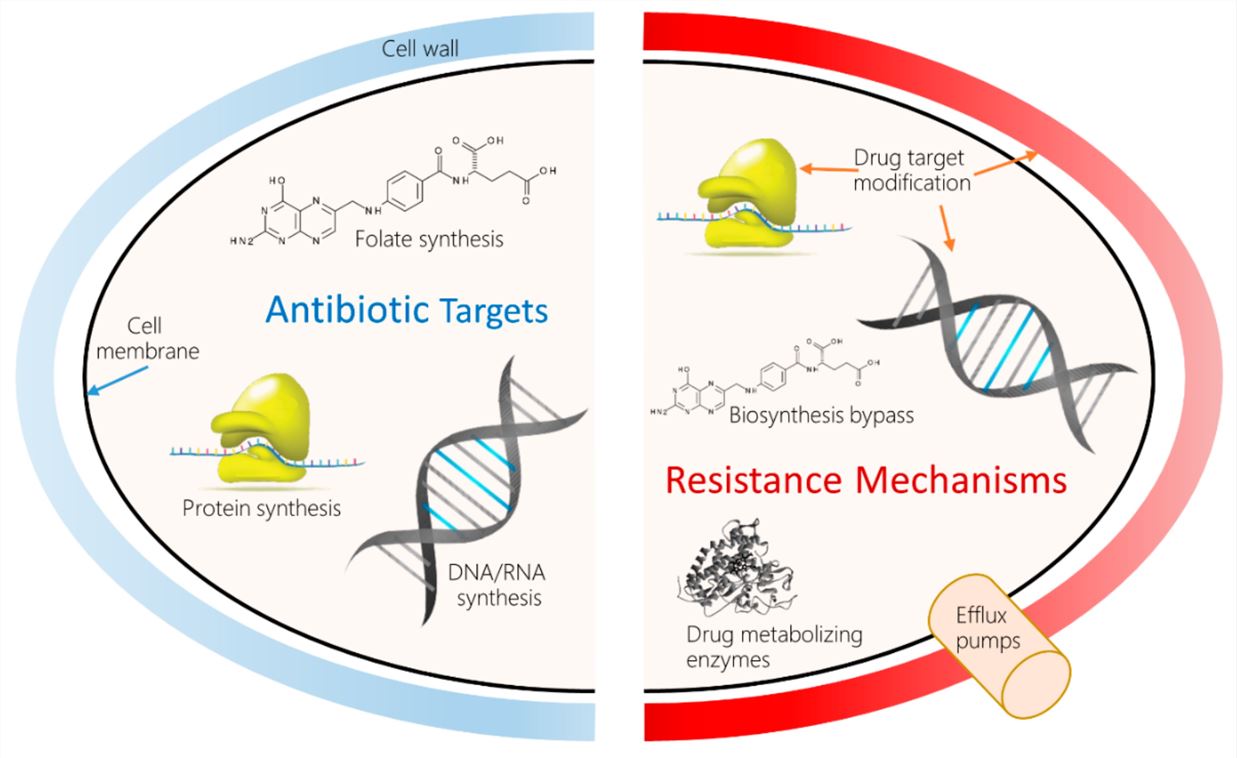 Fig.1 Antibacterial drug targets and molecular mechanisms of antibacterial drug resistance (AMR).1,3
Fig.1 Antibacterial drug targets and molecular mechanisms of antibacterial drug resistance (AMR).1,3
Available Antimicrobial Resistance (AMR) Models
By tailoring the specific characteristics of various drug-resistant bacteria, Creative Biolabs has established specialized AMR models to provide valuable tools for preclinical drug-resistant bacteria research and development.
| AMR Models | Related Disease & Drug Evaluation | Animal Species |
| SystemicCandidiasis Models | SystemicCandidiasis, Invasive Fungal Infections; for the evaluation of Antifungaldrugs (e.g.,Azoles, Echinocandins, Polyenes). | Mouse |
| PulmonaryInfection Models (General Pneumonia) | BacterialPneumonia (Healthcare-Associated Pneumonia, Hospital-Acquired Pneumonia, Community-AcquiredPneumonia); used for evaluating Antibiotics, Anti-virulence factors, andVaccines. | Mouse,Rat, Rabbit |
| PulmonaryInfection Models | P. aeruginosa Pneumonia, Ventilator-Associated Pneumonia (VAP); forevaluating Novel Antibiotics (especially against MDR strains), Anti-virulenceagents, and Phage therapy. | Mouse,Rabbit, Guinea Pig, NHPs |
| UrinaryTract Infection (UTI) Models | Uropathogenic E. coli (UPEC) Infection, Pyelonephritis; used for the evaluation of Antibiotics,Anti-adhesion therapies, and Vaccines. | Mouse,Rat, Guinea Pig |
| IntraperitonealInfection Models | Peritonitis,Intra-abdominal Sepsis, for evaluating broad-spectrum Antibiotics andAnti-fungals. | Mouse,Rat |
| SubcutaneousAbscess Models | SSTIs,MRSA infection, for evaluating Antibiotics, Anti-virulence agents, andTopical disinfectants. | Mouse,Rat, Rabbit |
| LymphNode Tuberculosis Models | Tuberculosis(TB), Extrapulmonary TB; used for the evaluation of Anti-Tuberculosis drugs (e.g., Isoniazid,Rifampicin) and Novel combination therapies. | Mouse,Guinea Pig, Rabbit |
| IntestinalColonization Models | GutDysbiosis, C.difficile Infection (CDI); for evaluating Probiotics/Prebiotics,Fecal Microbiota Transplantation (FMT), and targeted Antibiotics (e.g., Vancomycin). | Mouse,Rat |
| InvasivePulmonary Aspergillosis Models | InvasivePulmonary Aspergillosis (IPA) in immunocompromised hosts; used for theevaluation of Antifungal drugs and Immunomodulatory agents. | Mouse,Rat, Rabbit |
| Acute/ChronicInfection Models | AcuteSystemic Bacterial/fungal persistence Infections, Chronic Osteomyelitis,Chronic Wound Infections, biofilm formation; rapid efficacy screening ofnovel antibiotics, antivirals, and sepsis-targeting biologics. | Mouse,Rat, Rabbit, NHPs |
| In Vitro Drug Resistance Induction Experiment Models | Bacteria/Fungicultures in laboratory media; used for the development of next-generationAntibiotics and combination therapies. | Bacteria,Fungi |
| Non-Mammalian(Alternative) Infection Models | Pathogenicityscreening, rapid drug toxicity/efficacy, for initial screening of Antibioticsand Antifungals. | Zebrafish,Fruit fly |
Available Pathogens
The pathogens commonly employed in our established models include:
| Gram-positive drug-resistant bacteria | Gram-negative resistant bacteria |
| Methicillin-Resistant Staphylococcus aureus (MRSA) | Extended-Spectrum β-Lactamase-Producing Enterobacteriaceae (ESBL-E) |
| Vancomycin-Resistant Enterococcus (VRE) | Carbapenem-Resistant Enterobacteriaceae (CRE) |
| Penicillin-Resistant Streptococcus pneumoniae (PRSP) | Multidrug-Resistant Pseudomonas aeruginosa (MDR-PA) |
| Vancomycin-Resistant Staphylococcus aureus (VRSA) | Multidrug-Resistant/ Extensively Drug-Resistant Acinetobacter baumannii (MDR/XDR-AB) |
Measurements
We assessed the success of the modeling and its initial efficacy by measuring pathogen burden and evaluating organ damage. Our methods are specifically designed to quantitatively evaluate the effectiveness of novel therapeutic candidates against drug-resistant pathogens, ensuring scientific rigor and supporting robust decision-making in the drug development pipeline.
- Physiological status assessment: Continuous monitoring of physiological indicators such as body weight, temperature, activity levels, and respiratory rate is conducted following infection.
- Immune function analysis: Lymphocyte subset and pro-inflammatory factor levels are detected using flow cytometry, MSD, and ELISA.
- Pathogenic Detection: Accurate quantification (CFU/GE Quantification) of the pathogen load in multi-organ and tissue samples (such as infected tissue homogenates, blood, and urine) is performed through inoculation onto selective culture media and/or qPCR/RT-PCR techniques.
- Histopathological Analysis: H&E staining is used to assess the degree of inflammatory infiltration and tissue damage; simultaneously, special staining or immunohistochemistry (IHC) is utilized to identify the pathogen's location, biofilm structure, or immune cell subsets.
- Dynamic imaging technology: Employ fluorescently labeled drug-resistant bacteria, such as GFP-labeled MRSA, to observe the infection spread pathway in real time.
Applications
- Disease Modeling: AMR models simulate clinically challenging or chronic infections caused by resistant strains (e.g., biofilm formation) in animal hosts to replicate real-world pathology.
- Mechanism Study: AMR models, particularly through in vitro induction and alternative species models, allow for the rapid screening of compounds and the elucidation of the specific molecular mechanisms and evolutionary pathways of bacterial resistance.
- Drug Development: AMR models integrate in vitro PK/PD studies and in vivo infection challenges to precisely assess the efficacy of novel antibiotics and optimize dosing regimens against resistant pathogens.
- Anti-Infective Strategy Research: AMR models are used to evaluate and validate non-antibiotic strategies, such as anti-virulence drugs, phage therapy, or infection control measures, in combating resistance spread and infection.
Our Advantages
- Professional technology and experience: We have a professional scientific research team that integrates microbiologists, pharmacologists, and biostatisticians, offering in-depth services including mechanistic investigations and Pharmacology and Pharmacodynamic models. We possess expertise in selecting appropriate animal species and models to develop methodologies tailored to diverse research needs.
- Rich Model resources and customization capabilities: We offer an extensive repository of drug-resistant bacterial strains. This enables us to rapidly provide relevant models based on client specifications and develop customized animal models for specialized research needs.
- Cost and Efficiency Advantages: Leveraging large-scale and specialized operations, we optimize resource allocation in laboratory animal procurement and equipment utilization, thereby reducing the cost per project. Our extensive experience and mature technical processes facilitate the rapid establishment and execution of drug-resistant bacterial animal models, shortening project cycles, enhancing R&D efficiency, and accelerating experimental outcomes and drug development timelines for our clients.
- Stringent Quality Control and Management: A stringent quality control system has been implemented, with Standard Operating Procedures (SOPs) governing every stage from experimental animal sourcing and husbandry to bacterial inoculation, sample collection, and subsequent analysis. This meticulous oversight ensures the accuracy and reliability of all experimental results.
- Complex Model Development: We provide a comprehensive service chain, encompassing pre-experimental optimization, formal modeling, and efficacy evaluation, all customizable to client-specific needs.
Work with Us
- Summarize the project requirements and fill in the information collection form.
- Sign a CDA from both parties to further communicate information, such as targets.
- Select an animal model, discuss experimental design, and determine assay parameters.
- Project costing and project schedule forecasting.
- We provide a detailed project plan, including the required sample quantities, methods, and protocols.
- Both parties confirm the project details and start the project.
- Confirm the timeline of the project.
- We provide periodic results and information on the animal's condition.
- We will work together to make project adjustments as necessary.
- We provide a comprehensive project report promptly.
- We arrange transportation for the produced samples.
- We provide a discussion of the project results and help to arrange the next steps.
- Data storage and archiving.
FAQs
-
Q: What is the core value of our in vivo PD modeling services?
A: We utilize physiologically relevant animal infection models to generate highly clinically predictive PK/PD parameters. This approach ensures risk reduction in subsequent clinical trials.
-
Q: How do in vivo PD studies help us overcome AMR challenges?
A: In vivo models are the only platforms capable of fully assessing the complex interplay between the host immune system, drug penetration into the site of infection, and competition among resistant strains. By using animal models, we can determine the PD target, the minimum drug exposure required in a living system to overcome resistant bacteria, which guides rational dosing scheme design.
-
Q: Which animal model platforms do we primarily use for evaluation?
A: We employ multiple validated infection models, including but not limited to the neutropenic mouse thigh/pneumonia models, UTI infection models, and sepsis models. These platforms cover critical infection sites like the bloodstream, lungs, and urinary tract, allowing for flexible matching to the specific characteristics of different antimicrobials and resistant pathogens.
-
Q: How do our in vivo PD services accelerate the new drug development process?
A: By providing early and quantitative insights into the drug's in vivo PK/PD relationship and PD targets (e.g., AUC/MIC), we help clients rapidly optimize dosing regimens and filter lead candidates. This significantly shortens the R&D cycle and optimizes resource allocation.
-
Q: How do we use animal models to assess the risk of resistance emergence?
A: We design drug gradient exposure studies in models to monitor the frequency of resistance mutations in the bacterial population post-treatment. This in vivo assessment helps validate the Mutant Prevention Concentration (MPC) concept and provides crucial data on the drug's capacity to effectively suppress resistance emergence.
Published Data
Evaluation of in vivo efficacy and drug resistance of phage-antibiotic synergistic therapy (PAS) in a mouse model: The bacterial load in the cecum, kidneys, lungs, spleen, and the drug resistance rate of the PAS group were significantly lower than that of the single-drug groups, demonstrating that the combination can inhibit the emergence of drug resistance.
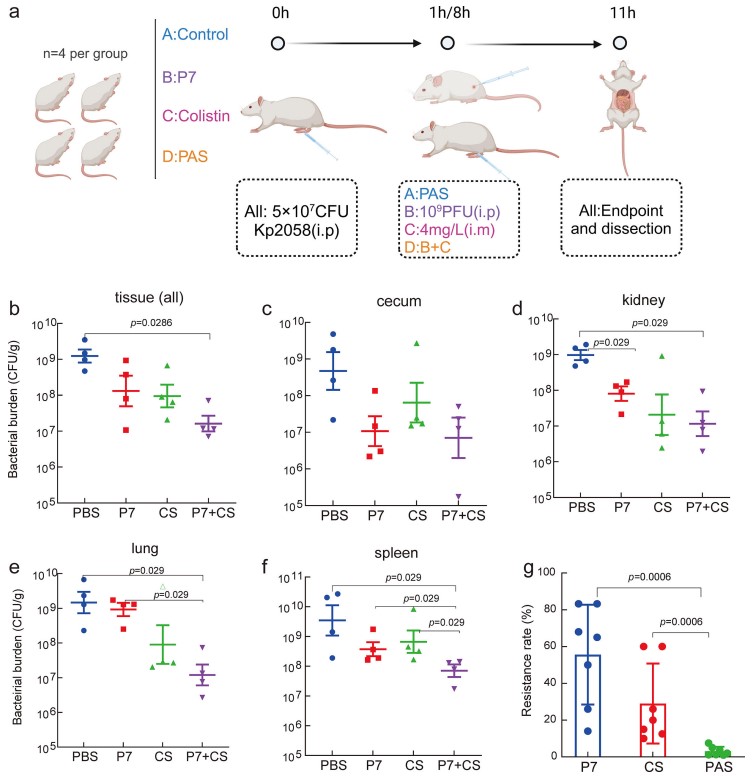 Fig.2 In vivo assessment of phage-antibiotic combination therapy against K. pneumoniae infection.2,3
Fig.2 In vivo assessment of phage-antibiotic combination therapy against K. pneumoniae infection.2,3
References
- Van Camp, Pieter-Jan et al. "Bioinformatics Approaches to the Understanding of Molecular Mechanisms in Antimicrobial Resistance." International Journal of molecular sciences vol. 21,4 1363. https://doi.org/10.3390/ijms21041363
- Zhao, Mengshi et al. "Antibacterial effect of phage cocktails and phage-antibiotic synergy against pathogenic Klebsiella pneumoniae." mSystems vol. 9,9 (2024): e0060724. https://doi.org/10.1128/msystems.00607-24
- Distributed under Open Access license CC BY 4.0, without modification.
For Research Use Only.
