B-Cell Epitope Prediction
Identifying the B-cell epitope is an important process in antibody discovery and also the key to effective antibody design. Serving as a leader and supplier in the biochemical market, Creative Biolabs has developed an original web-based immunogenicity screening platform to capture properties of potential antibodies to predict the safety and efficacy of these antibody targets. We believe, with our team of experts and most up-to-date facilities, we could help you to tailor the perfect B-cell epitope to suit the best to your projects.
Introduction to B-cell Epitope Prediction
B-cells are an important part of the immune system, as they enable trigger long-time immune responses against various viruses and pathogens. In this process, antibodies can recognize and bind with specific antigens (epitopes). B-cell epitopes are usually composed of two types, linear or conformational B-cell epitopes. Linear B-cell epitopes are continuous amino acid sequence fragments. Conformational B-cell epitopes consist of the amino acid that is mainly made up of different residues formed by protein folding.
Recently, pilot studies have shown that the epitope identification for T and B cells is critical to the development of high-affinity monoclonal antibodies (mAbs) against various human diseases. The experimental and computer-aid epitope identification has aroused much attention in the last several decades. Until now, many experimental assays have been generated for identifying B-cell epitopes, such as X-ray crystallography, enzyme-linked immunosorbent assays (ELISA), as well as peptide-chip. However, data have indicated that these methods are expensive and time-consuming with low throughput or accuracy. As a result, several computational approaches have been designed to assist or replace the experimental methods.
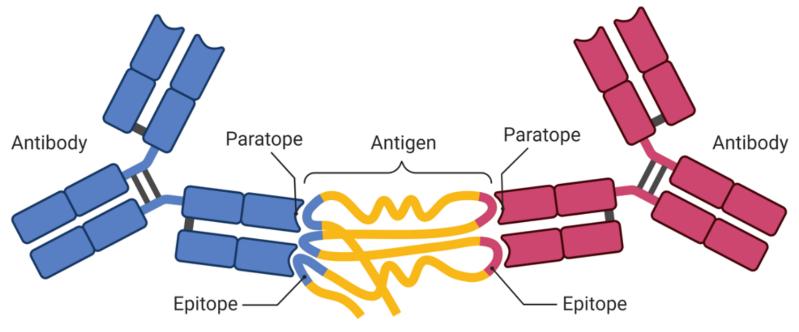 Fig.1 Antigen recognition by antibodies.1, 2
Fig.1 Antigen recognition by antibodies.1, 2
Our Services for B-cell Epitope Prediction
Epitope identification is a significant step in the development of effective antibodies for treating a range of human diseases. Up to now, B-cell epitope prediction tools have been divided into two main types: protein sequence-and structure-based approaches. The results derived from many studies have suggested that structure-based assays represent better performance in comparison to sequence-based assays. Besides, the definition of suitable datasets plays an essential role in predicting B-cell epitopes of your interest.
 Fig.2 Linear and conformational B-cell epitopes.1, 2
Fig.2 Linear and conformational B-cell epitopes.1, 2
At Creative Biolabs, we have developed a comprehensive B-cell epitope prediction platform to analyze the immunogenicity properties of potential antibodies, forecast their safety and effectiveness in pre-clinical and clinical use, and so much more. Currently, we offer a wide variety of B-cell epitope prediction services by using sequence-based, residue-based, and structure-based methods. Our in silico assays for linear B-cell epitope mapping are based on assessing a battery of physicochemical properties, including flexibility, hydrophilicity, and solvent solubility. A score to each residue of protein sequence will be assigned, and residues that have high scores may belong to B-cell epitopes.
Also, some conformational B-cell epitope prediction tools have been established to analyze the primary or the tertiary structure of candidate antibodies, as well as the interfacial residues involved in antibody complex formation.
The Protocol for B-cell Epitopes Prediction
- Paste the amino acid sequence into online B-Cell Epitope prediction tools
- Choose the prediction method
- Select the length of the epitope
- Pick the specificity of the classifier
- Submit a query to obtain predicted epitopes
Creative Biolabs has developed a novel web-based antibody immunogenicity assessment platform to remove the obstacles of your projects. Our scientists specialized in epitope prediction will work with you to develop a most appropriate strategy that will offer meaningful data for your research, please feel free to contact us for closer communication to learn how we can be involved in your project. Separate services or integrated end-to-end solutions are all welcomed.
References
- Bukhari, Syed Nisar Hussain, et al. "Machine learning techniques for the prediction of B-cell and T-cell epitopes as potential vaccine targets with a specific focus on SARS-CoV-2 pathogen: A review." Pathogens 11.2 (2022): 146.
- Under Open Access license CC BY 4.0, without modification.
For Research Use Only.
