In Vitro ADME Assays: Principles, Applications & Protocols
Introduction of ADME Assays
Overview of In Vitro ADME Testing
In the realm of pharmaceutical research, ADME (Absorption, Distribution, Metabolism, Excretion) assays serve as fundamental tools for evaluating the pharmacokinetic properties of drug candidates. These in vitro tests mimic physiological processes to predict how compounds behave in vivo, focusing on four key phases: how a drug enters the bloodstream (absorption), distributes to tissues (distribution), is metabolized (metabolism), and is eliminated from the body (excretion). By characterizing these properties early in drug development, researchers can identify candidates with favorable drug-like attributes, minimize risks in clinical trials, reduce development costs, and streamline the path to regulatory approval.
Why In Vitro ADME Matters
The significance of in vitro ADME testing lies in its ability to flag compounds with poor bioavailability or toxicity at an early stage, preventing resource-intensive investments in flawed candidates. Additionally, regulatory bodies such as the FDA and EMA require robust ADME data for Investigational New Drug (IND) submissions, making these assays indispensable for compliance. By integrating in vitro insights, pharmaceutical teams can align their candidates with safety and efficacy benchmarks, enhancing the likelihood of translational success.
Core Principles of In Vitro ADME Assays
Summary of ADME Processes
Drug absorption, distribution, metabolism, and excretion (ADME) studies are crucial in drug development. Absorption evaluates how compounds cross biological barriers, often using Caco-2 cells or PAMPA models. Distribution investigates drug binding to plasma proteins and accumulation in tissues, measuring the unbound active drug fraction through methods like equilibrium dialysis. Metabolism primarily focuses on the liver, employing liver microsomes and hepatocytes to assess metabolic stability and potential drug-drug interactions. Excretion explores renal and biliary clearance mechanisms, involving models with OATs/OCTs transporters and biliary excretion assays. These ADME studies collectively assess drug bioavailability and in vivo behavior, predicting efficacy and safety.
Table 1: Key Characteristics of ADME Assays
| Process | Key Concepts | Experimental Models/Methods | Primary Goals |
|---|---|---|---|
| Absorption | Solubility, permeability, intestinal barrier | Caco-2 cells, MDCK cells, PAMPA assays | Predict intestinal absorption and passive diffusion |
| Distribution | Plasma protein binding, tissue affinity | Equilibrium dialysis, red blood cell partitioning | Measure unbound drug fractions and tissue uptake |
| Metabolism | Hepatic stability, enzyme interactions | Liver microsomes, hepatocytes, CYP450 inhibition assays | Assess metabolic clearance and drug-drug interaction risks |
| Excretion | Renal/biliary clearance, transporters | OAT/OCT transporter models, in vitro biliary excretion assays | Evaluate elimination pathways and transport mechanisms |
Service you may interested in
Applications in Drug Development
Lead Optimization
High-throughput screening (HTS) platforms enable rapid assessment of metabolic stability and permeability across large compound libraries. By prioritizing candidates with favorable ADME profiles, researchers can eliminate "non-drug-like" molecules early, accelerating the lead optimization cycle. For example, compounds with short half-lives in liver microsome assays may be reformulated or discarded to avoid poor in vivo performance.
Toxicity Prediction
In vitro assays play a critical role in safety profiling. hERG potassium channel inhibition assays, for instance, identify compounds with potential to cause cardiac arrhythmias, a major safety concern. Metabolite identification using techniques like liquid chromatography-mass spectrometry (LC-MS/MS) helps detect reactive intermediates that may lead to toxicity, allowing for structural modifications to mitigate risks.
Clinical Translation
In vitro data are used to predict human pharmacokinetic parameters such as bioavailability, volume of distribution, and half-life. Physiologically based pharmacokinetic (PBPK) modeling integrates these insights to simulate in vivo behavior, supporting IND submissions with data-driven predictions of clinical performance. Validated in vitro assays provide regulatory agencies with confidence in a candidate's safety and PK profile.
Specialized Use Cases
For central nervous system (CNS) drug development, models like MDR1-MDCK cells (engineered to express efflux pumps like P-glycoprotein) assess blood-brain barrier (BBB) permeability, a critical criterion for CNS penetrance. In prodrug optimization, in vitro assays evaluate enzymatic activation in target tissues, ensuring that inactive precursors are efficiently converted to their active forms.
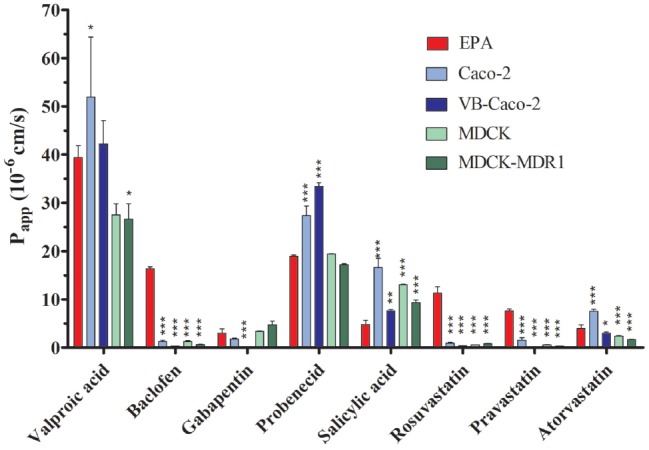 Fig.1 Comparison of Drug Permeability Across BBB and Epithelial Cell Models. 1,2
Fig.1 Comparison of Drug Permeability Across BBB and Epithelial Cell Models. 1,2
Standard Protocols for Key ADME Assays
Metabolic Stability
Liver Microsome Assay:
Incubate the test compound with human or rat liver microsomes, co-factors (e.g., NADPH), and buffer at 37°C.
Sample at predefined time points and quantify parent compound degradation via LC-MS/MS to calculate half-life and clearance rates.
Hepatocyte Stability Protocol:
Use cryopreserved human hepatocytes to mimic in vivo hepatic metabolism.
Measure intrinsic clearance (CLint) by monitoring substrate depletion over time, providing insights into first-pass metabolism and hepatic extraction.
Permeability Testing
Caco-2 Cell Monolayer Protocol:
Culture Caco-2 cells on transwell inserts to form a polarized monolayer (simulating intestinal epithelium).
Measure apical-to-basolateral (or basolateral-to-apical for efflux) transport of the compound using HPLC or MS, calculating permeability coefficients (Papp).
PAMPA Workflow:
Deploy artificial lipid membranes (e.g., phospholipid-coated filters) between donor and acceptor chambers.
Quantify compound passage over time to assess passive diffusion, particularly useful for high-throughput screening of lipophilicity.
Plasma Protein Binding
Equilibrium Dialysis:
Separate bound and unbound drug fractions using dialysis membranes with molecular weight cutoffs.
Quantify both fractions via LC-MS/MS to determine the free fraction (fu), a key parameter for predicting drug distribution and efficacy.
CYP450 Inhibition
Use fluorescent substrates (e.g., 7-ethoxyresorufin for CYP1A2) or LC-MS-based assays to measure enzyme activity in the presence of test compounds.
Determine IC50 values to assess inhibition potency for key isoforms (e.g., CYP3A4, CYP2D6), guiding DDI risk assessments.
Challenges and Optimization Strategies
Common Pitfalls
- Data Variability: Batch-to-batch differences in cell lines, microsome preparations, or reagent quality can lead to inconsistent results. Standardizing protocols and using validated reference compounds (e.g., verapamil for permeability assays) helps mitigate this.
- Low Throughput: Traditional manual assays are time-consuming, limiting their use in HTS. Transitioning to automated platforms addresses this bottleneck.
Technological Innovations
- Automation: Robotic liquid handlers and integrated workflows (from sample preparation to data analysis) enhance precision and throughput, enabling parallel processing of hundreds of compounds.
- Machine Learning (ML): ML models trained on curated ADME datasets predict properties like CLint, solubility, and permeability, reducing the need for extensive wet-lab testing.
- Advanced Analytics: Techniques like RapidFire-MS and ultra-performance liquid chromatography (UPLC) enable rapid metabolite profiling, improving the speed and resolution of metabolic stability assays.
Future Directions
- Organ-on-a-Chip Models: These microphysiological systems, which mimic organ-specific architectures (e.g., liver or intestine-on-a-chip), offer more physiologically relevant ADME predictions than traditional cell cultures, particularly for complex interactions involving multiple cell types.
- Multi-Omics Integration: Combining metabolomics, proteomics, and transcriptomics data provides a holistic view of drug effects, enabling more accurate in vitro-to-in vivo extrapolation (IVIVE).
- Regulatory Harmonization: Efforts to standardize IVIVE frameworks and validation criteria will enhance the reproducibility of in vitro data and streamline regulatory reviews.
In vitro ADME assays are indispensable pillars of modern drug development, providing critical insights into compound behavior that guide candidate selection, optimize lead structures, and mitigate clinical risks. As technology advances—through automation, machine learning, and innovative models like organ-on-a-chip—the field is poised to achieve even greater predictive power and efficiency. By embracing validated protocols, leveraging technological innovations, and fostering regulatory alignment, the pharmaceutical industry can accelerate the discovery of safe and effective therapies, ensuring that in vitro ADME remains at the forefront of translational science.
If you want to learn more about the transgenic mice, please refer to:
- Comprehensive Guide to In Vitro ADME Studies in Drug Discovery
- In Vitro ADME Screening: Accelerating Drug Development
- Differences and Connections Between ADME and DMPK in Drug Research
- In Vitro ADME Profiling Services: Accelerating Drug Development Through Predictive Insight
- In Vitro ADME and In Vivo PK Studies: A Holistic Framework for Drug Evaluation
- ADME in Toxicology: Ensuring Drug Safety and Efficacy
References
- Veszelka, Szilvia, et al. "Comparison of a rat primary cell-based blood-brain barrier model with epithelial and brain endothelial cell lines: Gene expression and drug transport." Frontiers in molecular neuroscience 11 (2018): 166. https://doi.org/10.3389/fnmol.2018.00166
- Distributed under Open Access license CC BY 4.0, without modification.
For Research Use Only.
